Technical Platforms
- Superresolution light microscopy facility
- Genetic engineering facility
- Mass spectrometry
- Confocal Laser Scanning Microscopy
- Life Cell Imaging
- PRECISE Platform foR SinglE Cell GenomIcS and Epigenomics
- Aptamer development platform at CARD
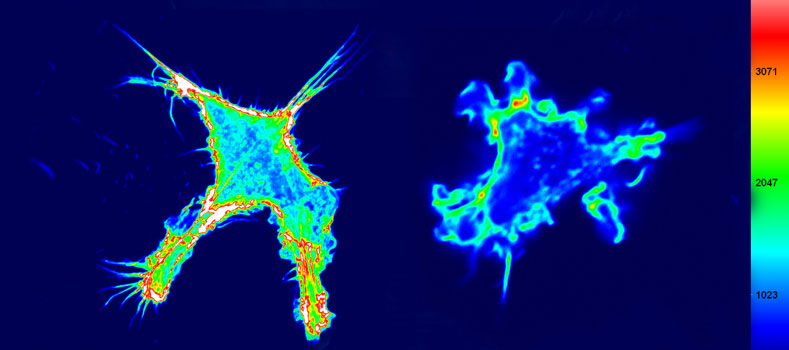
In life sciences, light microscopy is commonly used for the visualization of cellular structures. However, diffraction of light limits the resolution of microscopes to 250 nm. This resolution is not sufficient to resolve cellular details such as dense filaments, crowds of small objects or membranes forming an organelle.
STED (stimulated emission depletion) microscopy uses features of fluorescent dyes for circumventing the diffraction limit. The method has been invented and realized in the nineties and further improved and made commercially available at the beginning of this millennium. For his work on STED microscopy, Stefan Hell has been awarded with the Nobel Prize in chemistry in 2014.
Since summer 2015, in the LIMES ‘Superresolution light microscopy’ facility a 4-channel easy3D superresolution STED microscope (Abberior Instruments) is available allowing for imaging at resolutions down to 30 nm. In contrast to electron microscopy, which traditionally is used for studying cellular details at high resolution, the method does not require sample embedding, cutting into thin slices, and imaging in the vacuum. Even live cells can be studied. Moreover, specific labelling of proteins can be achieved easier than in electron microscopy.
Cells are the smallest units of life. Understanding their architecture and components is crucial for the further development of life science. STED microscopy allows studying a variety of cellular details as nanodomains, organelles or cytoskeleton and thereby helps unravelling the molecular complexity of living matter.
Contact: Prof. Dr. Thorsten Lang, thorsten.lang@uni-bonn.de
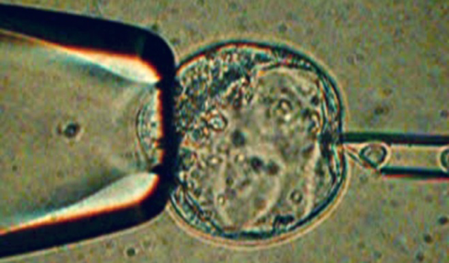
The development of CRISPR/Cas9 in the last decade has greatly advanced the manipulation of the genome in embryonic stem cells or fertilized eggs. It very much accelerated the process of such projects and enabled us to try much more complex changes. At the LIMES Institute, we routinely perform such experiments using Drosophila melanogaster, Xenopus tropicalis, zebra fish, as well as mouse models. The Genome Editing Core facility is your first contact from project support to taking over of whole projects.
Contact: Dr. Stephan Sonntag, stephan.sonntag@uni-bonn.de
Mass spectrometry is the key analytical technology for identification and quantitation of proteins, lipids, carbohydrates, small metabolites and pharmacologically active substances. LIMES owns a Thermo Q Exactive Plus that is mainly used for lipidomics and metabolomics, instrumental for research on lipid metabolism, metabolic diseases and their relation to the immune system. Its precision of typically 0.001 Da allows identification of substances just by their mass, while its resolution of 280000 allows separation of mass peaks that differ by not more than 0.002 Da.
Contact: Prof. Dr. Christoph Thiele, cthiele@uni-bonn.de

The Confocal Laser Microscopy platform provides state-of-the-art confocal imaging technology for cells and tissues along with scientific and technical expertise to support users in experimental design, image acquisition and analyses. The platform is based on several standard as well as advanced confocal systems which allow both high-resolution and ultra-fast imaging. Co-localization studies, fluorescent recovery after photobleaching (FRAP), fluorescence resonance energy transfer (FRET) can be conducted as well as imaging of photoactivatable fluorescent proteins. Additional processing software and workstations are available for post-acquisition analyses (2D, 3D and 4D).
Contact: Dr. Thomas Quast, quast@uni-bonn.de
The Live Cell Imaging platform offers an advanced microscopical equipment to monitor both intracellular dynamics or cell motility. We have long-standing expertise in the microscopical analysis of cell adhesion, migration and chemotaxis and we developed numerous assays to image dynamic processes with high spatial and temporal resolution. Recently, we established a microfluidic assay to analyze immune cell migration with high spatio-temporal resolution in precision controlled, visualized chemokine gradients. The platform offers highly spezialized equipment for automated and sample-preserving video-microscopy, including contrast enhancing, wide-field epifluorescence and confocal imaging systems. We also provide scientific and technical support for users to design experiments and to learn handling.
Contact: Dr. Thomas Quast, quast@uni-bonn.de
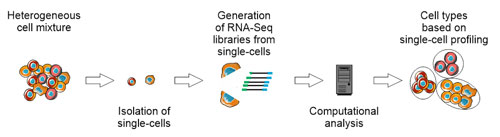
With the development of single-cell RNA sequencing, the research field of transcriptome analysis and system biology has been revolutionized. Until now the identification of single cells was based only on a limited number of predefined markers using existing methods such as FACS, single-cell RNA sequencing can be used to characterize cells on the basis of several hundred to thousands of genes. For example, the use of this technology has led to the discovery of previously unknown neurons (Zeisel et al., Science, 2015) and immune cells (Villani et al., Science, 2017).
To further promote the introduction of single-cell technologies into the laboratory routine, the “Platform for Single Cell Genomics and Epigenomics” was launched. PRECISE is a joint venture between the University of Bonn and the German Center for Neurodegenerative Diseases (DZNE). One particular focus is to develop and to apply genomic technologies to the single-cell level and also making them available to internal as well as external cooperation partners.
Within PRECISE the following single-cell techniques are used:
- MARS-Seq: MARS-Seq (Jaitin et al., Science, 2014) is an automated highly-parallelized single-cell sequencing method. It is based on the isolation of single-cells using FACS with subsequent RNA sequencing. Since the information of the FACS is stored, it can later be linked to the transcriptome information. Since the information of FACS are stored, they can be linked afterwards with the transcriptome information.
- SMART-Seq2: The SMART-Seq2 method (Picelli et al., Nature Protocols, 2013) uses the SMART reaction (switching mechanism at the end of the 5'-end of the RNA transcript). The precursor of this method was already developed in 2013 (Picelli et al., Nature Methods, 2013). While MARS-Seq sequences only the 3'-end, SMART-Seq2 allows sequencing of the entire transcript.
- Drop-Seq: Since single-cell technologies are often better and more reproducible in nanolite reactions, Drop-Seq (Macosko et al., Cell, 2015) has been developed, which makes use of microfluidics. With Drop-Seq, thousands of single-cells can be sequenced in a short time and at low cost.
- SeqWell: The SeqWell system offers an alternative to Drop-Seq (Gierahn et al., Nature Methods, 2017). As well as in the case of Drop-Seq, the reactions are also carried out in the nanoliter volume. In contrast to Drop-Seq, however, this is done in microwells combined with a semi-permeable membrane. One major advantage of the SeqWell is the portability of this technology.
- C1: The C1 Platform is similar to Drop-Seq and is based on microfluidics. It is distributed by Fluidigm. A special feature of this technology is the identification of single-cells in specific "capture sites", where they can be visualized. PRECISE uses C1 to perform SMART-Seq.
Contact: Prof. Dr. Joachim L. Schultze, Office.immunogenomics@uni-bonn.de
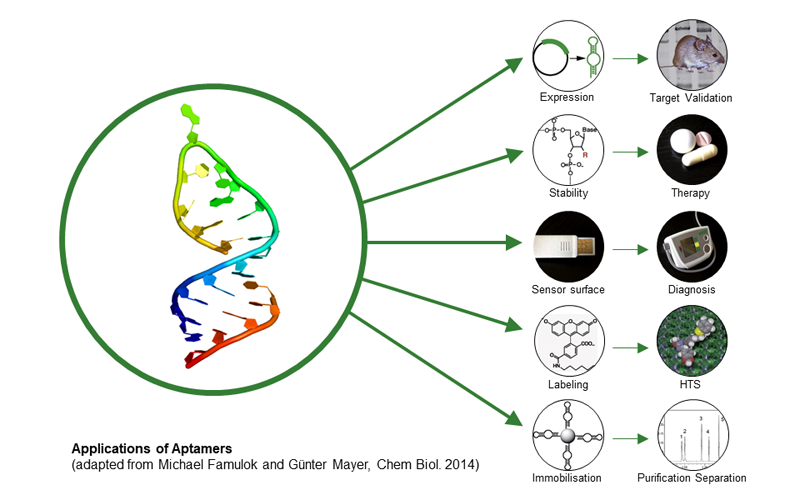
Aptamers are small single-stranded nucleic acids that fold into 3D structure and are able to bind various target molecules with high affinity and specificity similar to antibodies. Aptamers are generated by using an in vitro selection technique (SELEX), which is based on the amplification of the nucleic acid with the highest affinity. Based on this technique, aptamers can be generated against toxins, peptides, proteins, viruses, bacteria, and even whole cells. By introducing modified nucleotide building blocks, nuclease resistant aptamers can be generated and applied to different therapeutic areas. The broad applicability of aptamers makes them an universal biomolecule and an excellent alternative to existing antibodies.
Since January 2017, the Center of Aptamer Research and Development (CARD) was established at the University of Bonn. The technical center and its aptamer selection services are open to everyone, from academia to biotech or pharma companies. The following services are available: generation of new aptamers by using an automated in vitro selection process, an annual workshop on aptamer selection, theoretical and practical support in the field of aptamer application and research. For further information please visit our website www.aptamer-center.de.
Contact:
Prof. Dr. Günter Mayer, gmayer@uni-bonn.de